Multimedia Journal of Metaverse in MEDICINE
RESEARCH ARTICLE | JANUARY 2, 2025
NAT1 as a diagnostic and prognostic marker in colon cancer
Hao Chen1, Kang Li1, Kun Zhu1, Xin Wang1, Qiong Wang1, Chengxue Dang1, Yong Zhang1*
1 Department of Surgical Oncology, The First Affiliated Hospital of Xi’an JiaoTong University, Xi’an 710061, China.
Corresponding Authors: Yong Zhang. E-mail: [email protected]
Address: Department of Surgical Oncology, The First Affiliated Hospital of Xi’an JiaoTong University, Xi’an 710061, China.
Abstract
Background: Colorectal cancer is one of the most common malignant tumors of the digestive tract, and early diagnosis and intervention are of great significance. More diagnostic biomarkers need to be developed to improve the accuracy of colon cancer diagnosis. Method: The author explored the expression differences of NAT1 in tumor and healthy control tissues by downloading gene expression data from colorectal cancer patients and healthy controls in the TCGA database. The author also examined the expression of NAT1 protein using the HPA database。OC curves were used to ascertain the diagnostic performance of NAT1 in colorectal cancer patients. Additionally, the correlation between NAT1 expression and clinical indicators was explored. Survival analysis was conducted to understand whether NAT1 could serve as a prognostic marker for colorectal cancer, and nomograms were created combining clinical indicators. Finally, CIBERSORT was used to explore the correlation between NAT1 expression and immune cell infiltration. Results: The author found that NAT1 is significantly low expressed in colorectal cancer. The diagnostic efficacy of NAT1 for colorectal cancer was assessed through ROC curves, which showed that the area under the ROC curve was 0.900 (95% CI: 0.854-0.946). The expression of NAT1 was correlated with T stage, N stage, M stage, CEA levels, and lymphatic invasion. Survival analysis revealed that patients with lower NAT1 expression had poorer overall survival (HR: 0.53, 95%CI: 0.35-0.79, p=0.002), progress free interval (HR: 0.55, 95%CI: 0.39-0.79, p=0.001), and disease specific survival ((HR: 0.43, 95%CI: 0.26-0.73, p=0.002), and nomograms were created in conjunction with clinical indicators. Immune infiltration analysis revealed that NAT1 expression is positively correlated with Th2 cells, T helper cells, T cells, CD8 T cells, B cells, aDC, and Th17 cells, and negatively correlated with NK cell infiltration. Conclusion: NAT1 can serve as a diagnostic and prognostic biomarker for colorectal cancer, accompanied by good diagnostic efficacy.
Keywords: NAT1; colorectal cancer; diagnostic maker; prognostic maker
Introduction
Colorectal cancer, as one of the most common malignant tumors of the digestive tract, has seen an increasing incidence rate in recent years[1]. In 2019, there were 283,322 new cases and 92,784 deaths in China[2]. Improving and standardizing the diagnostic and treatment processes for colorectal cancer is of great importance. Currently, the diagnosis of colorectal cancer mainly relies on endoscopic examination and pathological biopsy. Research has investigated diagnostic markers for colorectal cancer, indicating that 100P, FOXO1, and LPAR1 may act as biomarkers with high diagnostic performance[3]. The MAGE-C gene has also been confirmed as a diagnostic marker for colorectal cancer and may serve as a therapeutic target for colorectal cancer[4]. N-acetyltransferase 1 is an acetyltransferase that, as a phase II drug-metabolizing enzyme, may participate in the metabolism and excretion of carcinogenic substances[5]. NAT1 is expressed in almost all tissues, and studies have indicated that NAT1 expression is associated with the progression of breast cancer[6]. Additionally, some studies have reported that NAT1 indeed promotes the upregulation of E-cadherin expression in colorectal cancer cells[7]. This study investigates the diagnostic and prognostic performance of NAT1 in colorectal cancer using bioinformatics methods.
Methods
1 Differential Gene Expression
To validate the expression of NAT1 in pan-cancer analysis, the authors downloaded gene expression data of 33 types of malignancies, including colorectal cancer, from the TCGA database. The expression differences of NAT1 between 33 tumor tissues and their corresponding normal tissues were compared. Additionally, NAT1 expression differences between colorectal cancer and normal intestinal mucosa were specifically compared. To explore whether NAT1 protein expression also differs between colorectal cancer and normal tissues, data on NAT1 expression in these tissues were retrieved from the HPA database.
2 Diagnostic Performance and Clinical Relevance
The diagnostic efficiency of NAT1 in colorectal cancer patients from the TCGA database was evaluated usi-ng ROC curves and the area under the curve (AUC).
NAT1 expression was analyzed across clinical subgroupsto investigate its association with common clinical indic-ators, including T stage, N stage, M stage, CEA levels, and lymphatic invasion.
3 Prognostic Indicators and Nomogram Constr-uction
To validate the prognostic value of NAT1 in colorectal cancer patients, survival data were downloaded from the TCGA database, and patients were divided into high-risk and low-risk groups based on NAT1 median expression levels. Differences in overall survival, progression-free survival, and cancer-specific survival between the two groups were compared. Additionally, a nomogram predicting patient survival was developed by integrating clinical staging and NAT1 expression.
4 Immune Infiltration
The CIBERSORT algorithm was utilized to assess the infiltration of 22 distinct immune cell types. Spearman correlation analysis revealed the association between NAT1 gene expression and infiltration of immune cells.
Results
NAT1 exhibited differential expression between normal tissues and 14 malignancies, including colorectal cancer, with low expression in colorectal cancer tissues (Figure 1A-B). Similar results were observed in the HPA database, where NAT1 protein was also downregulated in colorectal cancer tissues (Figure 1C-D). The ROC curve analysis showed that the AUC for NAT1 in diagnosing colorectal cancer was 0.900 (95% CI: 0.854–0.946) (Figure 2A). Clinical correlation analysis revealed lower NAT1 expression in T3 and T4 patients compared to T2, and significantly reduced expression in N1 and N2 stages compared to N0 (Figure 2B-2C). Patients with distant metastases exhibited lower NAT1 expression(Figure 2D), and NAT1 was downregulated in patients with CEA >5 compared to those with CEA ≤5 (Figure 2E). Patients with lymphatic invasion showed significantly lower NAT1 expression (Figure 2F).
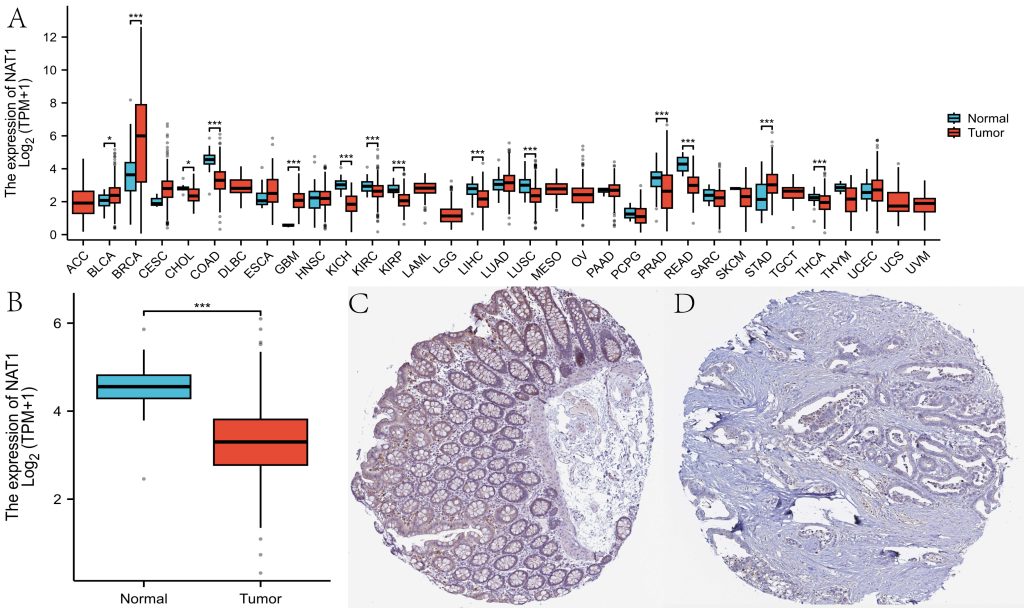
Fig.1 Differential Gene and Protein Expression.
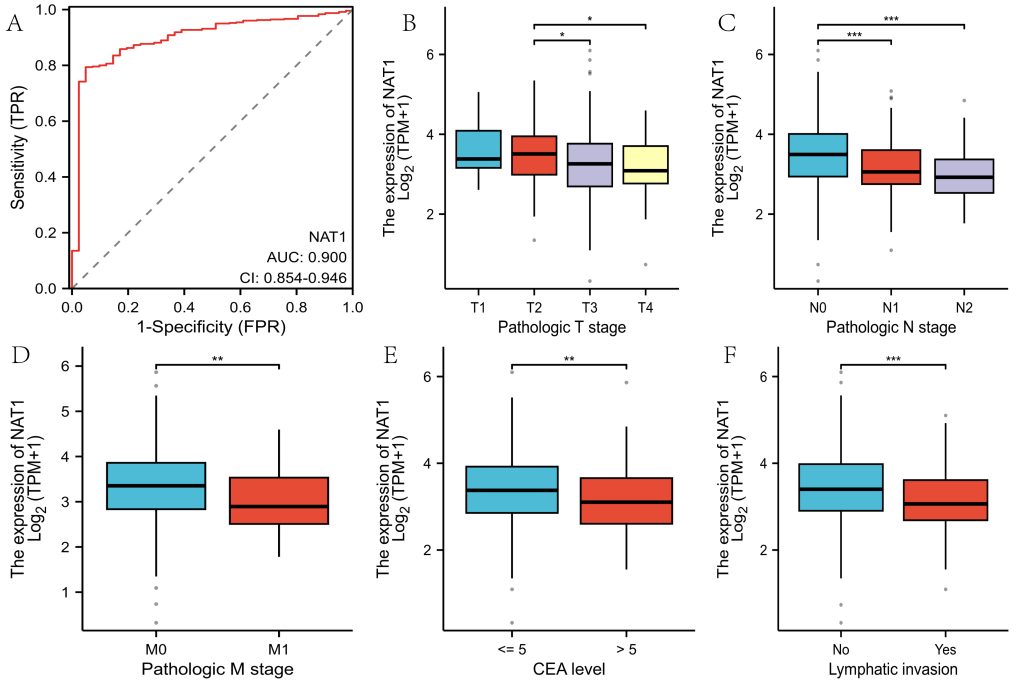
Fig.2 Diagnostic Performance and Clinical Relevance.
To investigate whether NAT1 expression is associated with patient survival outcomes, the authors extracted survival data from the TCGA database and divided patients into high-expression and low-expression groups. Survival analysis revealed that patients in the low-expression group had worse overall survival, progression-free interval, and disease-specific survival (Figure 3A-3C). Immune infiltration analysis indicated that NAT1 expression was positively correlated with Th2 cells, T helper cells, T cells, CD8 T cells, B cells, aDC, and Th17 cells, but negatively correlated with NK cell infiltration(Figure 4).
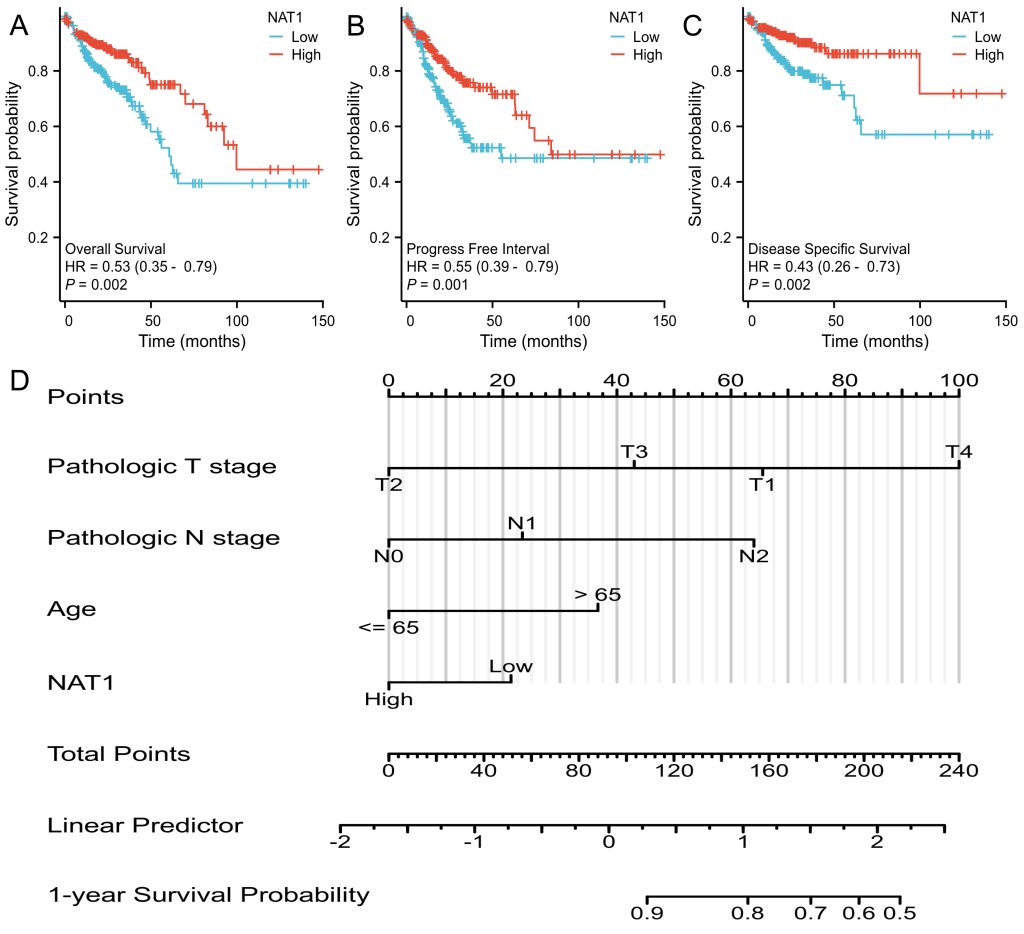
Fig.3 Prognostic Indicators and Nomogram Construction.
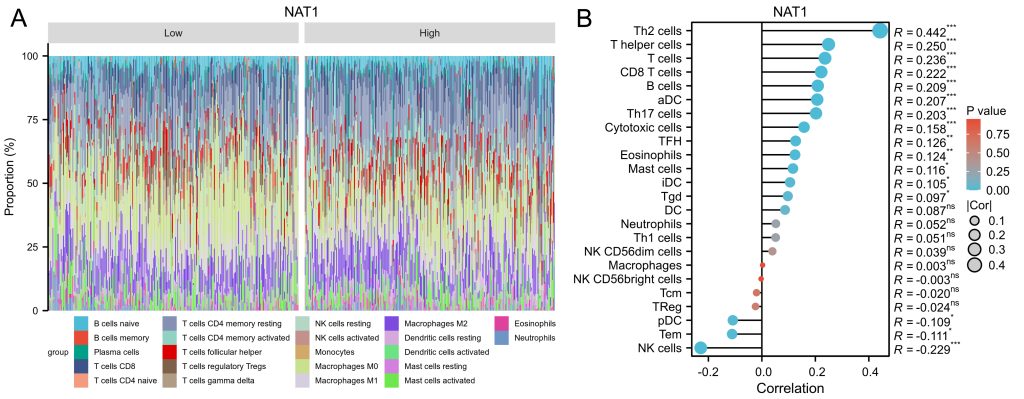
Fig.4 Immune Infiltration.
Discussion
The clinical symptoms of colorectal cancer are often atypical, making it easily confused with benign conditions such as hemorrhoids, which can delay treatment. Early screening, detection, and intervention can improve the 5-year survival rate of colorectal cancer patients, especially by detecting and removing adenomas at an early stage to prevent colorectal cancer. Currently, the diagnosis of colorectal cancer primarily relies on endoscopic examination and biopsy pathology. Reports suggest that some existing diagnostic biomarkers have suboptimal performance, highlighting the need for new diagnostic and prognostic markers. Some researchers have identified biomarkers such as GCNT2 using bioinformatics methods[8]. The author reviewed earlier literature to identify NAT1 as an acetyltransferase with a significant role in tumor metabolic reprogramming[9, 10]. Public database analysis showed NAT1 is downregulated in colorectal cancer tissues, with strong diagnostic utility and potential as a survival predictor. Additionally, immune infiltration analysis revealed that NAT1 expression was positively correlated with Th2 cells and T helper cells, and studies have shown that Th2 cells can inhibit colon cancer growth[11, 12]. NAT1 expression was negatively correlated with NK cells, and studies suggest that resting natural killer cells promote liver metastasis in colorectal cancer, consistent with our findings that patients with low NAT1 expression have a higher likelihood of liver metastasis. This study identified NAT1 as a diagnostic and prognostic biomarker for colorectal cancer using bioinformatics methods, and it is also associated with lymphatic and distant metastases in colorectal cancer patients. Limitations included biases of data from public databases and lacking confirmatory experiments demonstrating the expressions and functions of NAT1.
Conclusion
This study used bioinformatics methods to identify NAT1 as a diagnostic and prognostic biomarker for colorectal cancer, demonstrating strong diagnostic performance. The author intends to validate these results in cell lines and to investigate in animal models whether UGT2A3 can drive the progression of colitis-associated colon cancer.
Author Contributions
Hao Chen wrote the first draft. Kang Li, Kun Zhu, Xin Wang and Qiong Wang contributed to the critical revision of the manuscript. Chengxue Dang and Yong Zhang reviewed and revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by grants from Natural Science basic Research Project of Shaanxi Province (NO.2022JM-499).
Conflicts of Interest
The authors declare no conflict of interest.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Received: 22 November 2024
Accepted: 20 December 2024
Published on line: 2 January 2025
Reference
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin, 2024, 74: 12-49.
- Dong Y, Fan ZZ, Li WT et al. Burden of gastrointestinal cancers among working-age population over past thirty years in China. World J Gastrointest Oncol, 2024, 16: 3955-3979.
- Geng Y, Li Y, Liu G, Jiao J. Identification of biomarkers for the diagnosis in colorectal polyps and metabolic dysfunction-associated steatohepatitis (MASH) by bioinformatics analysis and machine learning. Sci Rep, 2024, 14: 29463.
- Almutairi MH, Alsoraie WA, Alrubie TM et al. Increased MAGE-C Family Gene Expression Levels as a Biomarker of Colon Cancer Through the Demethylation Mechanism. Pharmaceuticals (Basel), 2024, 17.
- Hickman D, Pope J, Patil SD et al. Expression of arylamine N-acetyltransferase in human intestine. Gut, 1998, 42: 402-409.
- Gupta RA, Motiwala MN, Dumore NG et al. Effect of piperine on inhibition of FFA induced TLR4 mediated inflammation and amelioration of acetic acid induced ulcerative colitis in mice. J Ethnopharmacol, 2015, 164: 239-246.
- Tiang JM, Butcher NJ, Cullinane C et al. RNAi-mediated knock-down of arylamine N-acetyltransferase-1 expression induces E-cadherin up-regulation and cell-cell contact growth inhibition. PLoS One, 2011, 6: e17031.
- Su Y, Tian X, Gao R et al. Colon cancer diagnosis and staging classification based on machine learning and bioinformatics analysis. Comput Biol Med, 2022, 145: 105409.
- Wise JTF, Salazar-Gonzalez RA, Walls KM et al. Hexavalent chromium increases the metabolism and genotoxicity of aromatic amine carcinogens 4-aminobiphenyl and beta-naphthylamine in immortalized human lung epithelial cells. Toxicol Appl Pharmacol, 2022, 449: 116095.
- Mohammadi H, Roochi MM, Sadeghi M et al. Association of N-acetyltransferases 1 and 2 Polymorphisms with Susceptibility to Head and Neck Cancers-A Meta-Analysis, Meta-Regression, and Trial Sequential Analysis. Medicina (Kaunas), 2021, 57.
- Liu JQ, Li XY, Yu HQ et al. Tumor-specific Th2 responses inhibit growth of CT26 colon-cancer cells in mice via converting intratumor regulatory T cells to Th9 cells. Sci Rep, 2015, 5: 10665.
- Jacenik D, Karagiannidis I, Beswick EJ. Th2 cells inhibit growth of colon and pancreas cancers by promoting anti-tumorigenic responses from macrophages and eosinophils. Br J Cancer, 2023, 128: 387-397.
